Description
Recognition Site:
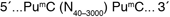
McrBC is an endonuclease which cleaves DNA containing methylcytosine* on one or both strands. McrBC will not act upon unmethylated DNA (1). Sites on the DNA recognized by McrBC consist of two half-sites of the form (G/A)mC. These half-sites can be separated by up to 3 kb, but the optimal separation is 55-103 base pairs (2,3). McrBC requires GTP for cleavage, but in the presence of a non-hydrolyzable analog of GTP, the enzyme will bind to methylated DNA specifically, without cleavage (4).
*5-methylcytosine or 5-hydroxymethylcytosine (5).
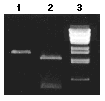 Linearized plasmid (methylated or unmethylated), containing one McrBC site, incubated with McrBC. Lane 1, unmethylated DNA; Lane 2, methylated DNA; Lane 3, Marker: Lambda DNA BstEII digest.
Linearized plasmid (methylated or unmethylated), containing one McrBC site, incubated with McrBC. Lane 1, unmethylated DNA; Lane 2, methylated DNA; Lane 3, Marker: Lambda DNA BstEII digest.
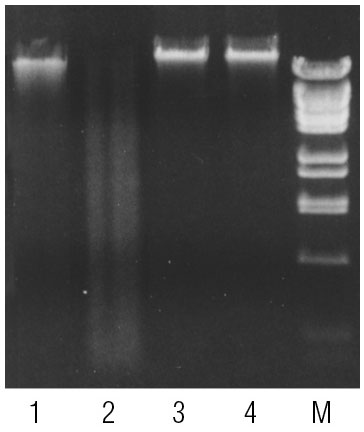 Human and Drosophila genomic DNA incubated with McrBC. 60-90% of CpGs in vertebrate DNA are estimated to be methylated (6) while methylated CpGs are extremely rare in Drosophila DNA (7). Lane 1, Human DNA; Lane 2, Human DNA incubated with McrBC; Lane 3, Drosophila DNA; Lane 4, Drosophila DNA incubated with McrBC; Lane 4, Marker: Lambda DNA BstEII digest.
Human and Drosophila genomic DNA incubated with McrBC. 60-90% of CpGs in vertebrate DNA are estimated to be methylated (6) while methylated CpGs are extremely rare in Drosophila DNA (7). Lane 1, Human DNA; Lane 2, Human DNA incubated with McrBC; Lane 3, Drosophila DNA; Lane 4, Drosophila DNA incubated with McrBC; Lane 4, Marker: Lambda DNA BstEII digest.
Product Source
The two component proteins are purified separately from E. coli K-12 strains containing plasmids encoding McrB and McrC (1).
- This product is related to the following categories: